De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis
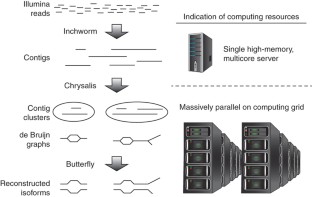
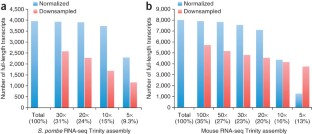
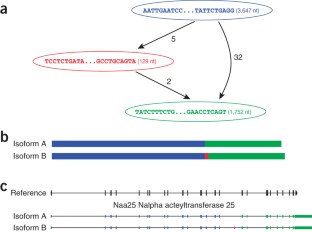
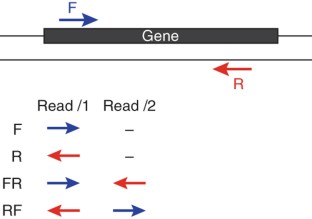
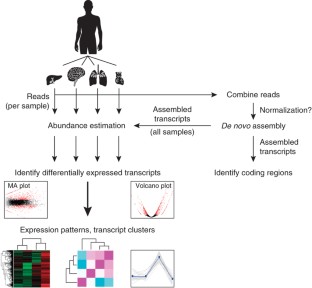
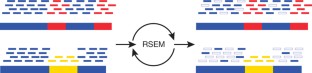
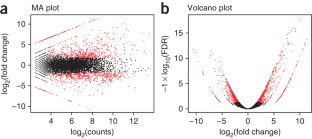
De novo assembly of RNA-seq data enables researchers to study transcriptomes without the need for a genome sequence; this approach can be usefully applied, for instance, in research on 'non-model organisms' of ecological and evolutionary importance, cancer samples or the microbiome. In this protocol we describe the use of the Trinity platform for de novo transcriptome assembly from RNA-seq data in non-model organisms. We also present Trinity-supported companion utilities for downstream applications, including RSEM for transcript abundance estimation, R/Bioconductor packages for identifying differentially expressed transcripts across samples and approaches to identify protein-coding genes. In the procedure, we provide a workflow for genome-independent transcriptome analysis leveraging the Trinity platform. The software, documentation and demonstrations are freely available from http://trinityrnaseq.sourceforge.net. The run time of this protocol is highly dependent on the size and complexity of data to be analyzed. The example data set analyzed in the procedure detailed herein can be processed in less than 5 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
265,23 € per year
only 22,10 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others

Partitioning RNAs by length improves transcriptome reconstruction from short-read RNA-seq data
Article 10 January 2022

Systematic assessment of long-read RNA-seq methods for transcript identification and quantification
Article Open access 07 June 2024
A Bayesian approach for accurate de novo transcriptome assembly
Article Open access 03 September 2021
References
- Wang, Z., Gerstein, M. & Snyder, M. RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet.10, 57–63 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Haas, B.J. & Zody, M.C. Advancing RNA-seq analysis. Nat. Biotechnol.28, 421–423 (2010). ArticleCASPubMedGoogle Scholar
- Martin, J.A. & Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet.12, 671–682 (2011). ArticleCASPubMedGoogle Scholar
- Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc.7, 562–578 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Guttman, M. et al. Ab initio reconstruction of cell type–specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol.28, 503–510 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Robertson, G. et al. De novo assembly and analysis of RNA-seq data. Nat. Methods7, 909–912 (2010). ArticleCASPubMedGoogle Scholar
- Schulz, M.H., Zerbino, D.R., Vingron, M. & Birney, E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics28, 1086–1092 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Grabherr, M.G. et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol.29, 644–652 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Duan, J., Xia, C., Zhao, G., Jia, J. & Kong, X. Optimizing de novo common wheat transcriptome assembly using short-read RNA-seq data. BMC Genomics13, 392 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Xu, D.L. et al. De novo assembly and characterization of the root transcriptome of Aegilops variabilis during an interaction with the cereal cyst nematode. BMC Genomics13, 133 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhao, Q.Y. et al. Optimizing de novo transcriptome assembly from short-read RNA-seq data: a comparative study. BMC Bioinformatics12 (suppl. 14), S2 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Henschel, R. et al. Trinity RNA-seq assembler performance optimization. XSEDE '12 Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: bridging from the eXtreme to the campus and beyond (Chicago, Illinois, USA, July 16–20, 2012) http://dx.doi.org/10.1145/2335755.2335842 (2012).
- Marcais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics27, 764–770 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Li, B. & Dewey, C.N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics12, 323 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics26, 139–140 (2010). ArticleCASPubMedGoogle Scholar
- Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol.11, R106 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Bullard, J.H., Purdom, E., Hansen, K.D. & Dudoit, S. Evaluation of statistical methods for normalization and differential expression in mRNA-seq experiments. BMC Bioinformatics11, 94 (2010). ArticlePubMedPubMed CentralCASGoogle Scholar
- Fang, Z. & Cui, X. Design and validation issues in RNA-seq experiments. Briefi. Bioinform.12, 280–287 (2011). ArticleCASGoogle Scholar
- Auer, P.L. & Doerge, R.W. Statistical design and analysis of RNA sequencing data. Genetics185, 405–416 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods5, 621–628 (2008). ArticleCASPubMedGoogle Scholar
- Trapnell, C. et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol.28, 511–515 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Roberts, A. & Pachter, L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods10, 71–73 (2013). ArticleCASPubMedGoogle Scholar
- Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol.10, R25 (2009). ArticlePubMedPubMed CentralCASGoogle Scholar
- Robinson, M.D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol.11, R25 (2010). ArticlePubMedPubMed CentralCASGoogle Scholar
- Dillies, M.A. et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform.http://dx.doi.org/10.1093/bib/bbs046 (17 September 2012).
- Marioni, J.C., Mason, C.E., Mane, S.M., Stephens, M. & Gilad, Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res.18, 1509–1517 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Robinson, J.T. et al. Integrative genomics viewer. Nat. Biotechnol.29, 24–26 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Abeel, T., Van Parys, T., Saeys, Y., Galagan, J. & Van de Peer, Y. GenomeView: a next-generation genome browser. Nucleic Acids Res.40, e12 (2012). ArticleCASPubMedGoogle Scholar
- Liu, L. et al. Comparison of next-generation sequencing systems. J. Biomed. Biotechnol.2012, 251364 (2012). PubMedPubMed CentralGoogle Scholar
- Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science323, 133–138 (2009). ArticleCASPubMedGoogle Scholar
- Rothberg, J.M. et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature475, 348–352 (2011). ArticleCASPubMedGoogle Scholar
- Van Belleghem, S.M., Roelofs, D., Van Houdt, J. & Hendrickx, F. De novo transcriptome assembly and SNP discovery in the wing polymorphic salt marsh beetle Pogonus chalceus (Coleoptera, Carabidae). PLoS ONE7, e42605 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kleinman, C.L. & Majewski, J. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science335, 1302 (2012). ArticleCASPubMedGoogle Scholar
- Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pounds, S.B., Gao, C.L. & Zhang, H. Empirical Bayesian selection of hypothesis testing procedures for analysis of sequence count expression data. Stat. Appl. Genet. Mol. Biol.http://dx.doi.org/10.1515/1544-6115.1773 (2012).
- Tarazona, S., Garcia-Alcalde, F., Dopazo, J., Ferrer, A. & Conesa, A. Differential expression in RNA-seq: a matter of depth. Genome Res.21, 2213–2223 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Cumbie, J.S. et al. GENE-counter: a computational pipeline for the analysis of RNA-seq data for gene expression differences. PLoS ONE6, e25279 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Hardcastle, T.J. & Kelly, K.A. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics11, 422 (2010). ArticlePubMedPubMed CentralGoogle Scholar
- Leng, N. et al. An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics29, 1035–1043 (2012). ArticleCASGoogle Scholar
- Tuna, M. & Amos, C.I. Genomic sequencing in cancer. Cancer Lett.http://dx.doi.org/doi:10.1016/j.canlet.2012.11.004 (2012).
- Rhind, N. et al. Comparative functional genomics of the fission yeasts. Science332, 930–936 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kumar, S. & Blaxter, M.L. Comparing de novo assemblers for 454 transcriptome data. BMC Genomics11, 571 (2010). ArticlePubMedPubMed CentralGoogle Scholar
- Papanicolaou, A., Stierli, R., Ffrench-Constant, R.H. & Heckel, D.G. Next generation transcriptomes for next generation genomes using est2assembly. BMC Bioinformatics10, 447 (2009). ArticlePubMedPubMed CentralCASGoogle Scholar
- Lohse, M. et al. RobiNA: a user-friendly, integrated software solution for RNA-seq–based transcriptomics. Nucleic Acids Res.40, W622–W627 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal17http://journal.embnet.org/index.php/embnetjournal/article/view/200/479 (2011).
- Haas, B.J., Chin, M., Nusbaum, C., Birren, B.W. & Livny, J. How deep is deep enough for RNA-seq profiling of bacterial transcriptomes? BMC Genomics13, 734 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Brown, C.T., Howe, A., Zhang, Q., Pryrkosz, A.B. & Brom, T.H. A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv:1203.4802 [q-bio.GN] (2012).
- Borodina, T., Adjaye, J. & Sultan, M. A strand-specific library preparation protocol for RNA sequencing. Methods Enzymol.500, 79–98 (2011). ArticleCASPubMedGoogle Scholar
- Parkhomchuk, D. et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res.37, e123 (2009). ArticlePubMedPubMed CentralCASGoogle Scholar
- Sung, W.K. et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet.44, 765–769 (2012). ArticleCASPubMedGoogle Scholar
Acknowledgements
We are grateful to D. Jaffe and S. Young for access to additional computing resources, to Z. Chen for help in R-scripting, to L. Gaffney for help with figure illustrations, to C. Titus Brown for essential discussions and inspiration related to digital normalization strategies, to G. Marcais and C. Kingsford for supporting the use of their Jellyfish software in Trinity and to B. Walenz for supporting our earlier use of Meryl. We are grateful to our users and their feedback, in particular J. Wortman and P. Bain for comments on earlier drafts of the manuscript. This project has been funded in part (B.J.H.) with Federal funds from the National Institute of Allergy and Infectious Diseases (NIAID), US National Institutes of Health (NIH), Department of Health and Human Services (DHHS), under contract no. HHSN272200900018C. Work was supported by Howard Hughes Medical Institute (HHMI), a NIH PIONEER award, a Center for Excellence in Genome Science grant no. 5P50HG006193-02 from the National Human Genome Research Institute (NHGRI) and the Klarman Cell Observatory at the Broad Institute (A.R.). A.P. was supported by the CSIRO Office of the Chief Executive (OCE). M.Y. was supported by the Clore Foundation. P.B. was supported by the National Science Foundation (NSF) grant no. OCI-1053575 for the Extreme Science and Engineering Discovery Environment (XSEDE) project. B.L. and C.D. were partially supported by NIH grant no.1R01HG005232-01A1. In addition, B.L. was partially funded by J. Thomson's MacArthur Professorship and by the Morgridge Institute for Research support for Computation and Informatics in Biology and Medicine. M.L. was supported by the Bundesministerium für Bildung und Forschung via the project 'NGSgoesHPC'. N.P. was funded by the Fund for Scientific Research, Flanders (Fonds Wetenschappelijk Onderzoek (FWO) Vlaanderen), Belgium. R.H. and R.D.L. were funded by the NSF under grant nos. ABI-1062432 and CNS-0521433 to Indiana University, and by Indiana METACyt Initiative, which is supported in part by Lilly Endowment, Inc. J.B. was supported through a CSIRO eResearch Accelerated Computing Project. Any opinions, findings and conclusions or recommendations expressed in this article are those of the authors and do not necessarily reflect the views of any of the funding bodies and institutions including the National Science Foundation, the National Center for Genome Analysis Support and Indiana University.
Author information
- Brian J Haas and Alexie Papanicolaou: These authors contributed equally to this work.
Authors and Affiliations
- Broad Institute of Massachusetts Institute of Technology (MIT) and Harvard, Cambridge, Massachusetts, USA Brian J Haas, Moran Yassour, Nathalie Pochet & Aviv Regev
- Commonwealth Scientific and Industrial Research Organisation (CSIRO) Ecosystem Sciences, Black Mountain Laboratories, Canberra, Australian Capital Territory, Australia Alexie Papanicolaou & Michael Ott
- The Selim and Rachel Benin School of Computer Science, The Hebrew University of Jerusalem, Jerusalem, Israel Moran Yassour & Nir Friedman
- Department of Medical Biochemistry and Microbiology, Science for Life Laboratory, Uppsala University, Uppsala, Sweden Manfred Grabherr
- Pittsburgh Supercomputing Center, Carnegie Mellon University, Pittsburgh, Pennsylvania, USA Philip D Blood
- CSIRO Information Management & Technology, St. Lucia, Queensland, Australia Joshua Bowden
- Department of Microbiology and Molecular Genetics, Oklahoma State University, Stillwater, Oklahoma, USA Matthew Brian Couger
- Genomics Research Centre, Griffith University, Gold Coast Campus, Gold Coast, Queensland, Australia David Eccles
- Department of Computer Sciences, University of Wisconsin, Madison, Wisconsin, USA Bo Li & Colin N Dewey
- Center for Information Services and High-performance Computing (ZIH), Technische Universität Dresden, Dresden, Germany Matthias Lieber
- California Institute for Quantitative Biosciences, University of California, Berkeley, Berkeley, California, USA Matthew D MacManes
- Institute for Genome Sciences, Baltimore, Maryland, USA Joshua Orvis
- Department of Plant Systems Biology, Department of Plant Biotechnology and Bioinformatics, Vlaams Instituut voor Biotechnologie (VIB), Ghent University, Ghent, Belgium Nathalie Pochet
- Parco Tecnologico Padano, Località Cascina Codazza, Lodi, Italy Francesco Strozzi
- United States Department of Agriculture–Agricultural Research Service, Corn Insects and Crop Genetics Research Unit, Ames, Iowa, USA Nathan Weeks
- Genomics facility, Purdue University, West Lafayette, Indiana, USA Rick Westerman
- GWT-TUD GmbH, Saxony, Germany Thomas William
- Department of Biostatistics and Medical Informatics, University of Wisconsin, Madison, Wisconsin, USA Colin N Dewey
- Research Technologies Division, University Information Technology Services, Indiana University, Bloomington, Indiana, USA Robert Henschel & Richard D LeDuc
- Department of Biology, Howard Hughes Medical Institute, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA Aviv Regev
- Brian J Haas














